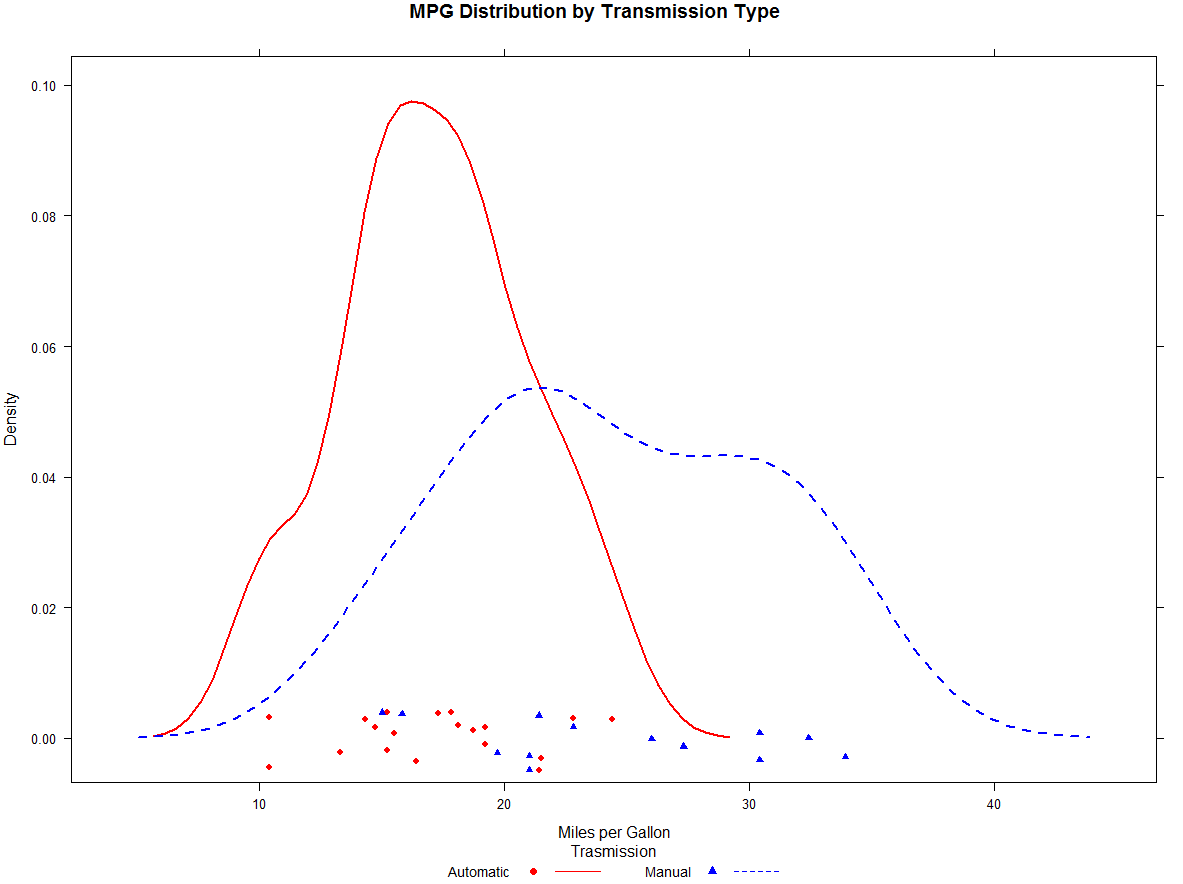
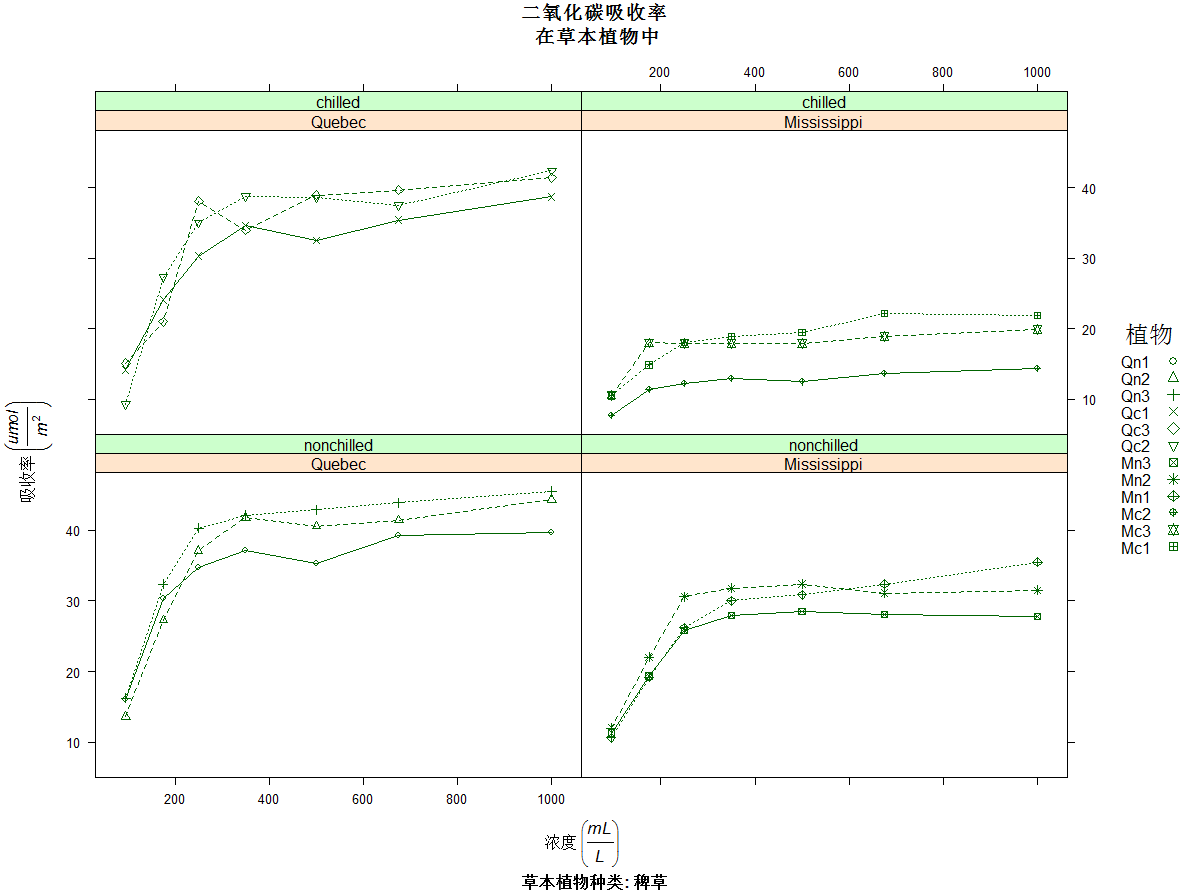
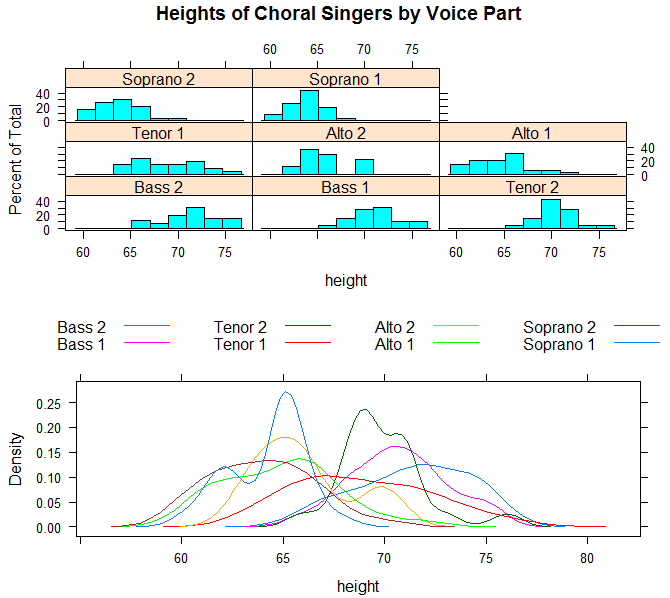
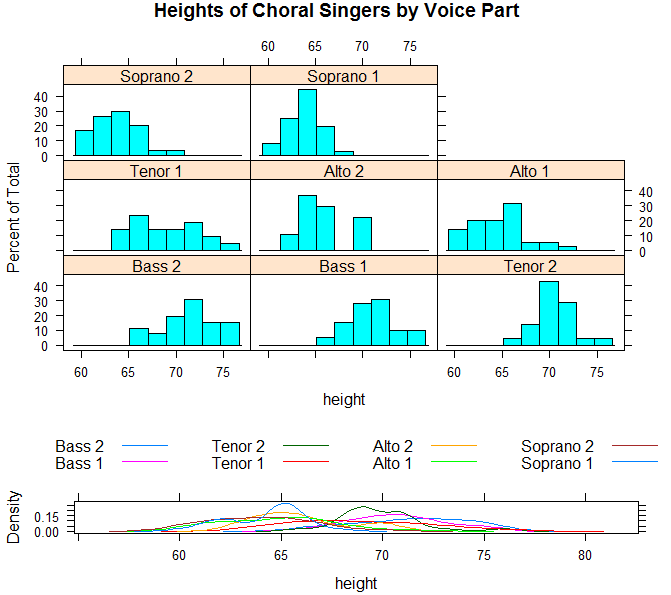
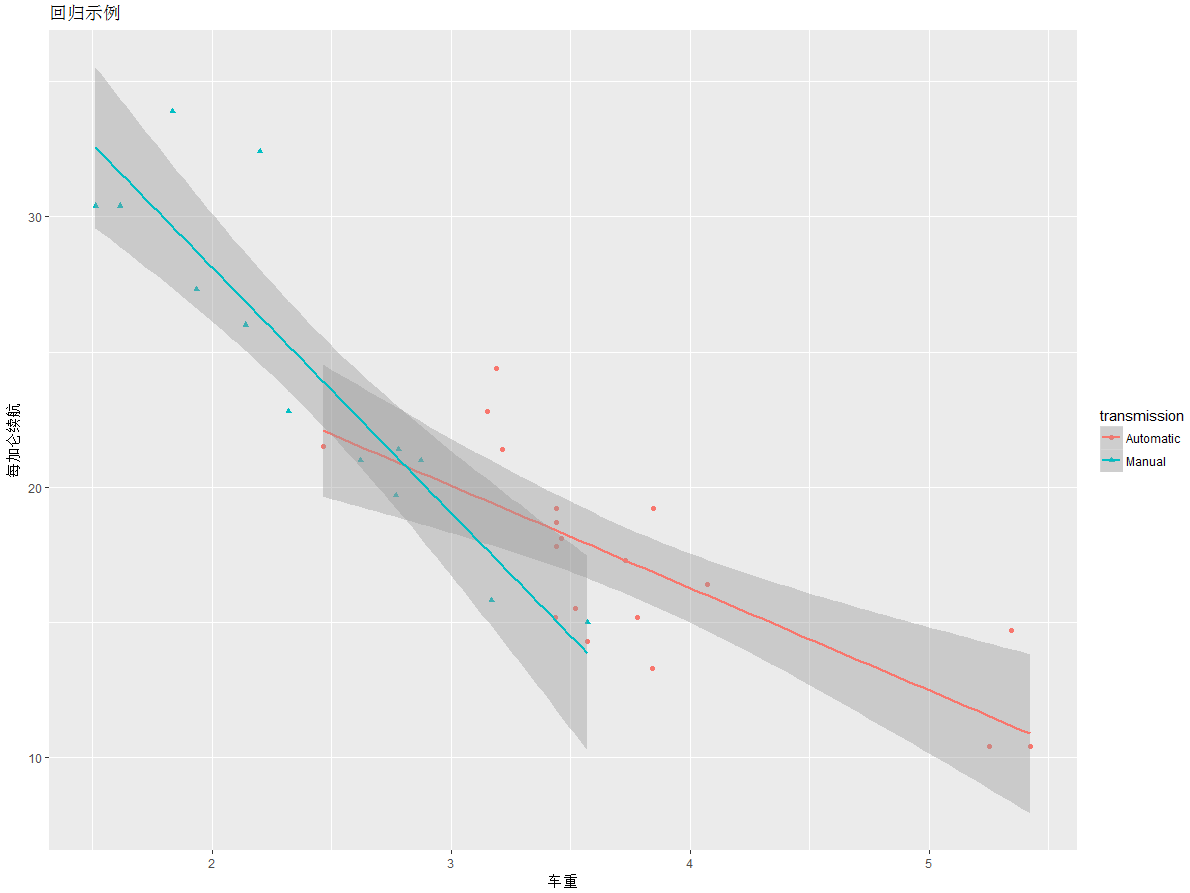
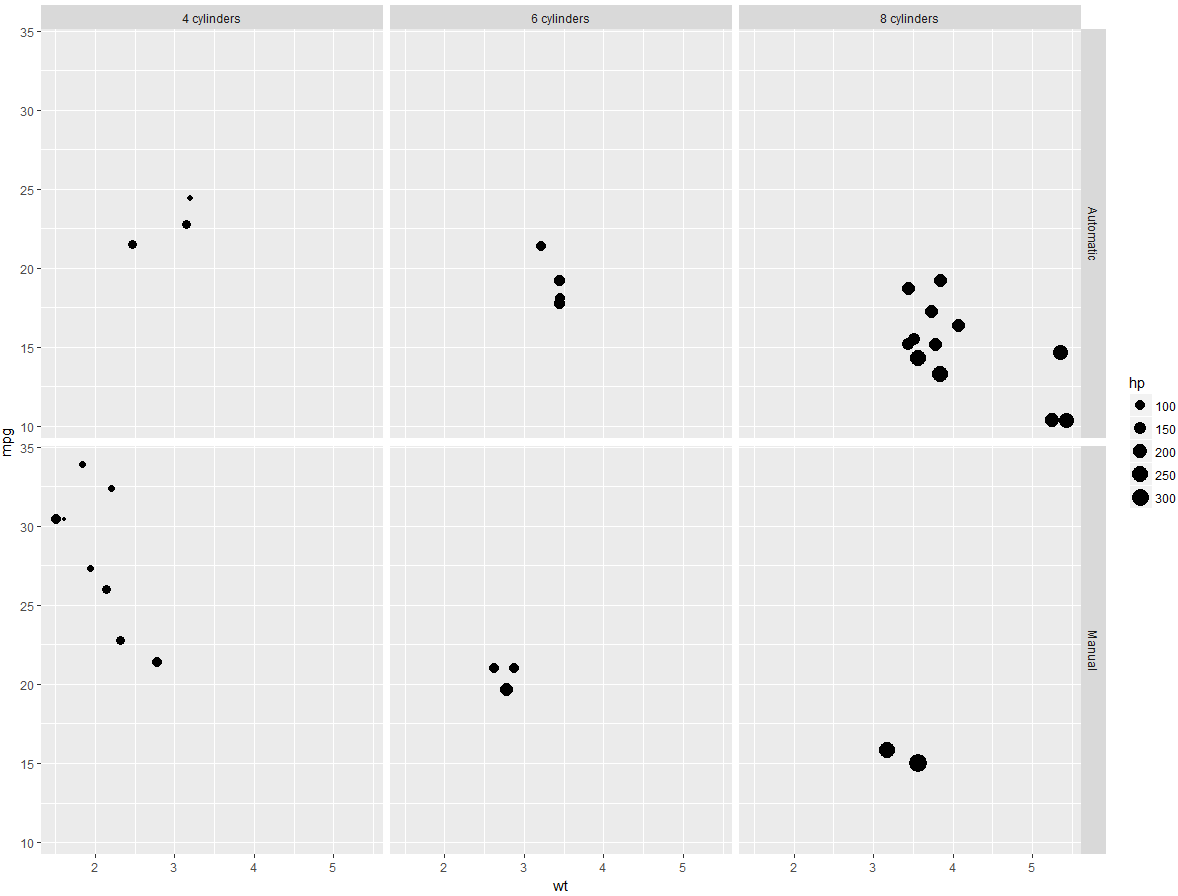
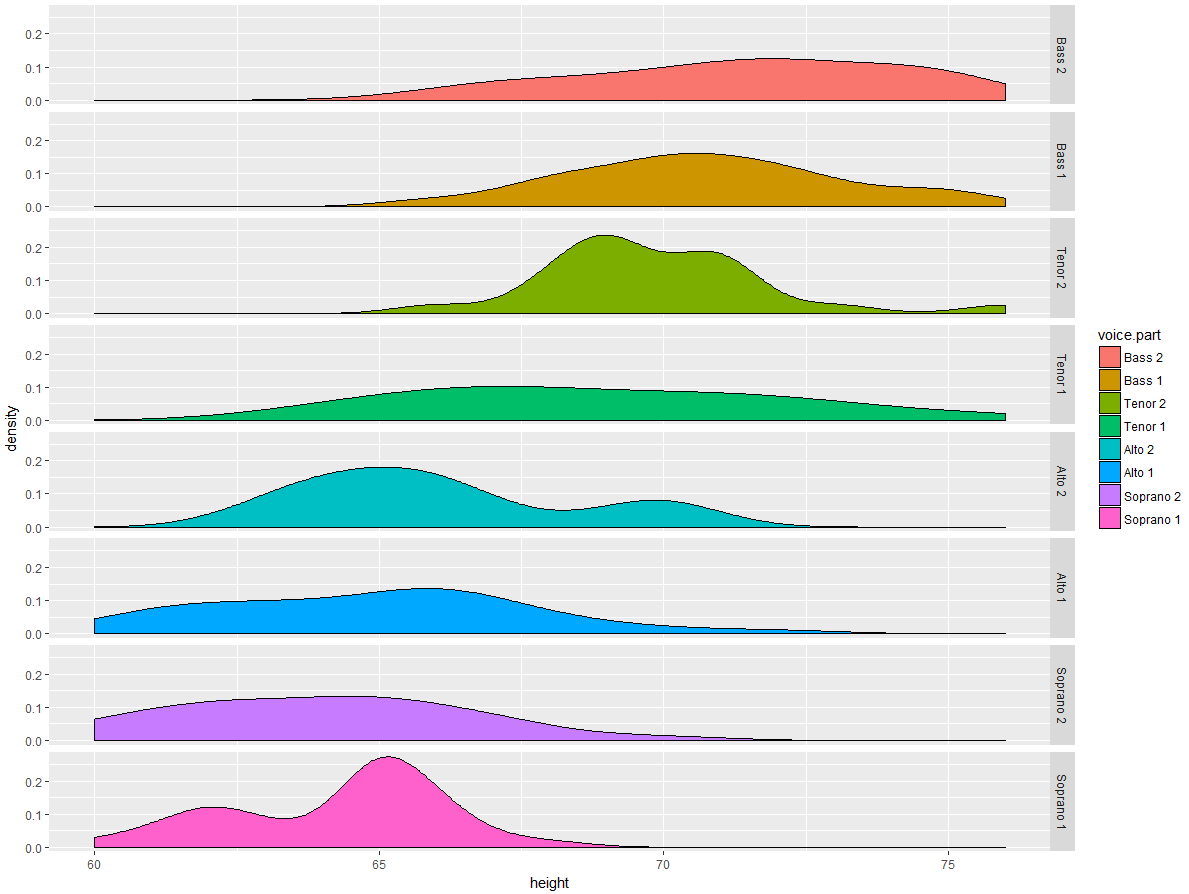
>
> library(mice)
>
>
>
>
>
>
>
>
>
> data(sleep, package="VIM")
> imp <- mice(sleep, seed=1234)
iter imp variable
1 1 NonD Dream Sleep Span Gest
1 2 NonD Dream Sleep Span Gest
1 3 NonD Dream Sleep Span Gest
1 4 NonD Dream Sleep Span Gest
1 5 NonD Dream Sleep Span Gest
2 1 NonD Dream Sleep Span Gest
2 2 NonD Dream Sleep Span Gest
2 3 NonD Dream Sleep Span Gest
2 4 NonD Dream Sleep Span Gest
2 5 NonD Dream Sleep Span Gest
3 1 NonD Dream Sleep Span Gest
3 2 NonD Dream Sleep Span Gest
3 3 NonD Dream Sleep Span Gest
3 4 NonD Dream Sleep Span Gest
3 5 NonD Dream Sleep Span Gest
4 1 NonD Dream Sleep Span Gest
4 2 NonD Dream Sleep Span Gest
4 3 NonD Dream Sleep Span Gest
4 4 NonD Dream Sleep Span Gest
4 5 NonD Dream Sleep Span Gest
5 1 NonD Dream Sleep Span Gest
5 2 NonD Dream Sleep Span Gest
5 3 NonD Dream Sleep Span Gest
5 4 NonD Dream Sleep Span Gest
5 5 NonD Dream Sleep Span Gest
>
> fit <- with(imp, lm(Dream ~ Span + Gest))
>
> pooled <- pool(fit)
>
> summary(pooled)
est se t df Pr(>|t|) lo 95 hi 95
(Intercept) 2.546199168 0.254689696 9.997260 52.12563 1.021405e-13 2.035156222 3.0572421151
Span -0.004548904 0.012039106 -0.377844 51.94538 7.070861e-01 -0.028707741 0.0196099340
Gest -0.003916211 0.001468788 -2.666287 55.55683 1.002562e-02 -0.006859066 -0.0009733567
nmis fmi lambda
(Intercept) NA 0.08710301 0.05273554
Span 4 0.08860195 0.05417409
Gest 4 0.05442170 0.02098354
>
>
> imp$imp$Dream
1 2 3 4 5
1 1.0 0.5 0.5 0.5 0.3
3 2.6 2.1 1.5 1.8 1.3
4 3.4 3.1 3.4 1.2 3.4
14 0.3 0.5 0.5 0.3 1.2
24 1.8 1.3 3.6 0.9 5.6
26 2.3 3.1 2.0 2.6 2.1
30 1.2 0.3 3.4 2.6 2.3
31 3.4 0.5 0.6 1.0 0.5
47 0.5 1.5 1.5 2.2 3.4
53 0.3 0.5 0.5 0.5 0.6
55 0.5 0.9 2.6 2.7 2.4
62 1.0 2.1 0.5 3.9 3.6
>
>
>
> complete(imp,action=1)
BodyWgt BrainWgt NonD Dream Sleep Span Gest Pred Exp Danger
1 6654.000 5712.00 3.3 1.0 3.3 38.6 645.0 3 5 3
2 1.000 6.60 6.3 2.0 8.3 4.5 42.0 3 1 3
3 3.385 44.50 9.7 2.6 12.5 14.0 60.0 1 1 1
4 0.920 5.70 13.2 3.4 16.5 4.7 25.0 5 2 3
5 2547.000 4603.00 2.1 1.8 3.9 69.0 624.0 3 5 4
6 10.550 179.50 9.1 0.7 9.8 27.0 180.0 4 4 4
7 0.023 0.30 15.8 3.9 19.7 19.0 35.0 1 1 1
8 160.000 169.00 5.2 1.0 6.2 30.4 392.0 4 5 4
9 3.300 25.60 10.9 3.6 14.5 28.0 63.0 1 2 1
10 52.160 440.00 8.3 1.4 9.7 50.0 230.0 1 1 1
11 0.425 6.40 11.0 1.5 12.5 7.0 112.0 5 4 4
12 465.000 423.00 3.2 0.7 3.9 30.0 281.0 5 5 5
13 0.550 2.40 7.6 2.7 10.3 7.0 52.0 2 1 2
14 187.100 419.00 3.2 0.3 3.1 40.0 365.0 5 5 5
15 0.075 1.20 6.3 2.1 8.4 3.5 42.0 1 1 1
16 3.000 25.00 8.6 0.0 8.6 50.0 28.0 2 2 2
17 0.785 3.50 6.6 4.1 10.7 6.0 42.0 2 2 2
18 0.200 5.00 9.5 1.2 10.7 10.4 120.0 2 2 2
19 1.410 17.50 4.8 1.3 6.1 34.0 252.0 1 2 1
20 60.000 81.00 12.0 6.1 18.1 7.0 12.0 1 1 1
21 529.000 680.00 11.9 0.3 12.5 28.0 400.0 5 5 5
22 27.660 115.00 3.3 0.5 3.8 20.0 148.0 5 5 5
23 0.120 1.00 11.0 3.4 14.4 3.9 16.0 3 1 2
24 207.000 406.00 10.4 1.8 12.0 39.3 252.0 1 4 1
25 85.000 325.00 4.7 1.5 6.2 41.0 310.0 1 3 1
26 36.330 119.50 10.9 2.3 13.0 16.2 63.0 1 1 1
27 0.101 4.00 10.4 3.4 13.8 9.0 28.0 5 1 3
28 1.040 5.50 7.4 0.8 8.2 7.6 68.0 5 3 4
29 521.000 655.00 2.1 0.8 2.9 46.0 336.0 5 5 5
30 100.000 157.00 9.7 1.2 10.8 22.4 100.0 1 1 1
31 35.000 56.00 10.4 3.4 13.8 16.3 33.0 3 5 4
32 0.005 0.14 7.7 1.4 9.1 2.6 21.5 5 2 4
33 0.010 0.25 17.9 2.0 19.9 24.0 50.0 1 1 1
34 62.000 1320.00 6.1 1.9 8.0 100.0 267.0 1 1 1
35 0.122 3.00 8.2 2.4 10.6 14.0 30.0 2 1 1
36 1.350 8.10 8.4 2.8 11.2 2.6 45.0 3 1 3
37 0.023 0.40 11.9 1.3 13.2 3.2 19.0 4 1 3
38 0.048 0.33 10.8 2.0 12.8 2.0 30.0 4 1 3
39 1.700 6.30 13.8 5.6 19.4 5.0 12.0 2 1 1
40 3.500 10.80 14.3 3.1 17.4 6.5 120.0 2 1 1
41 250.000 490.00 4.7 1.0 6.2 23.6 440.0 5 5 5
42 0.480 15.50 15.2 1.8 17.0 12.0 140.0 2 2 2
43 10.000 115.00 10.0 0.9 10.9 20.2 170.0 4 4 4
44 1.620 11.40 11.9 1.8 13.7 13.0 17.0 2 1 2
45 192.000 180.00 6.5 1.9 8.4 27.0 115.0 4 4 4
46 2.500 12.10 7.5 0.9 8.4 18.0 31.0 5 5 5
47 4.288 39.20 12.0 0.5 12.5 13.7 63.0 2 2 2
48 0.280 1.90 10.6 2.6 13.2 4.7 21.0 3 1 3
49 4.235 50.40 7.4 2.4 9.8 9.8 52.0 1 1 1
50 6.800 179.00 8.4 1.2 9.6 29.0 164.0 2 3 2
51 0.750 12.30 5.7 0.9 6.6 7.0 225.0 2 2 2
52 3.600 21.00 4.9 0.5 5.4 6.0 225.0 3 2 3
53 14.830 98.20 3.3 0.3 2.6 17.0 150.0 5 5 5
54 55.500 175.00 3.2 0.6 3.8 20.0 151.0 5 5 5
55 1.400 12.50 11.0 0.5 11.0 12.7 90.0 2 2 2
56 0.060 1.00 8.1 2.2 10.3 3.5 120.0 3 1 2
57 0.900 2.60 11.0 2.3 13.3 4.5 60.0 2 1 2
58 2.000 12.30 4.9 0.5 5.4 7.5 200.0 3 1 3
59 0.104 2.50 13.2 2.6 15.8 2.3 46.0 3 2 2
60 4.190 58.00 9.7 0.6 10.3 24.0 210.0 4 3 4
61 3.500 3.90 12.8 6.6 19.4 3.0 14.0 2 1 1
62 4.050 17.00 12.0 1.0 12.8 13.0 38.0 3 1 1